A Gemba Walk for Data Science
An excerpt from Altdeep.ai’s workshop Modern Probabilistic Modeler.
The following is excerpt from a non-technical part of Altdeep.ai’s workshop Modern Probabilistic Modeler.
Technical vs. Biological Variation
Consider a metabolic engineer who seeks to increase a microbes ability to convert a cheap and available source of carbohydrates into a vanillin, a valuable food additive.
She does so by introducing variations to the genomes of the microbe, creating an wide array of mutant strains. The strains are then colonized, then inserted in media that they can ferment. Technical measurement devices then quantify the concentration of vanillin, and strains that increase production relative to the original parent strain are preserved for the next iteration.
Let's tell the story:
Introduce the genetic variation using recombinant DNA technology.
Cultivate new strains on growth media.
Once the strains grow, cultivate them on fermentation media.
Take samples of the fermentation media and get measure the passage of light through the media.
Convert those light measurements to quantifications of the concentration of vanillin.
The goal of this data generating process is to identify which strain produces the most vanillin. However, we have to account for biological variation. If you take one batch of cultivated strain, split it up into twelve samples that each ferment the same fermentation media, you will get ultimately get 12 different concentrations of vanillin. That biological variation is intrinsic to the data generating process; microbiology is very stochastic, genetically identical cells in identical conditions given identical stimuli, will have slight differences in behavior.
Note that as probabilistic modelers, we will want to explicitly model intrinsic sources of stochasticity, such as biological variation in our models. Using probability to model intrinsic stochasticity in the data generating process is not the same as using probability to model our uncertainty about the data generating process. We'll cover this more later.
The engineer will want to average over each sample and compare the average. However, she also faces a second challenge of distinguishing biological variation from technical variation.
Technical variation is variation that occurs as a result of the process of collecting data. It contributes to variation in the data, just like biological variation. However, biological variation is actually interesting and something we want to understand. Technical variation gets in the way of that understanding.
Consider some sources of technical variation that could get in the way.
Growth and fermentation process
Creating growth media, fermentation media, and the growth and fermentation process itself are all processes were all kinds of technical variations can enter. Everything from changes in suppliers for the ingredients to the media to lighting conditions can influence the outcome.
Containers and transference devices
Containers and devices for transferring fluids between containers are primary sources of technical variation. Are there surface artifacts or foreign matter in the containers? What about air bubbles? Could they be contaminated? Could minerals like salt and calcium be accumulating on the inside, affecting precision?
Measurement Devices and Quantification
There are many technologies that can quantify concentration of a compound in a solution, each with different quirks. Most need regular servicing and calibration. Further, these devices never just magically return a vanillin concentration, the engineer needs to sort of process for converting raw measurements to concentrations, such as a calibration curve.
Methods for converting raw measurements to concentrations typically add nonlinear technical noise. For example, you may get unreliable data when the concentrations are too low or too high.
Generalizing: Modeling data collection noise vs. signal of interest
The microbiology laboratory example above is an excellent analogy for all data generating processes. There is going to be signal from the underlying phenomenon that you wish to model and understand, and there will be noise from processes that have nothing to do with that phenomena but still greatly impact the data.
You model should capture both.
Get up and personal with your noise
This manner of thinking is rare in modern data science practice, and especially rare in the machine learning community. How often do people in computer vision ask how the images were generated? On what cameras? Using what kind of film? What was the intent of the photographer? How were the images processed for storage on the server?
Management philosophy has approach called a gemba walk (related genchi genbutsu) which encourages managers to walk the physical spaces of operations (a factory floor, an office space, a physical environment) getting physically and technically familiar with the steps of the processes they are charged with managing.
The manager does gemba walks to form a mental model of the process so that they can use to make better decisions. How much more important that you, the probabilistic modeler, who will create an actual modeling artifact in code, should also do gemba walks?
Suppose you work in a company that builds and delivers a software application, and you are tasked with analyzing user behavior. Then the gemba walk in this case might come in the form of a user interview.
We are modeling the data generating process. That process ends when you import a data file into RStudio or a Jupyter notebook. Was that file created by a SQL query? What processes inserted the data into those tables? What trigger the insertion of that into the database? A probabilistic modeler needs to be as intimate with the data generating process as they can feasibly manage.
Only then can they understand sources of "technical variation" and account for it in the model.


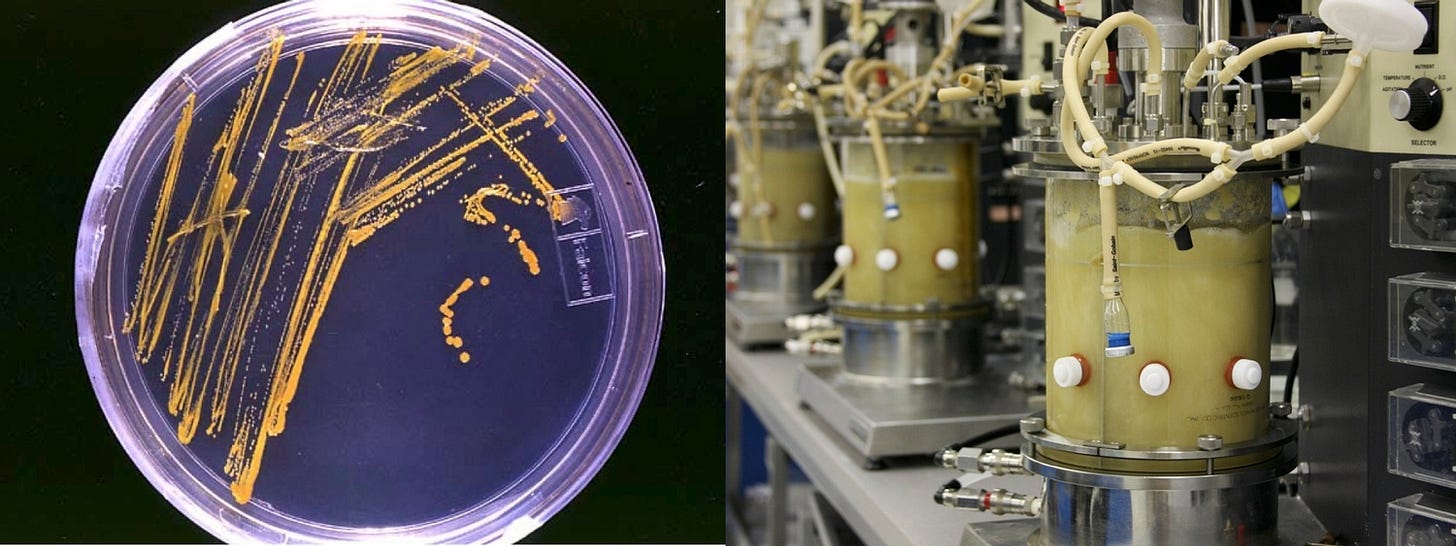




Never knew my time spent doing wet lab experiments was a gemba walk! I agree strongly I always recommend that computational biologists spend a little bit of time at the bench. I didn't just learn things about the process but also about the data.
For example the fact that for certain kinds of data , we choose a datapoint in the middle of the saturation curve that we then record. If I modeling the data and I have no idea that this data point came from a saturation curve I'm losing a lot of context